Author: Rory Tarnow-Mordi
In the first of this series of articles on Databricks, we looked at how Databricks works and the general benefits it brings to organisations ready to do more with their data assets. In this post we build upon this theme in the advanced analytics space. We will also walk through an interesting (biometric data generated by an Apple Watch) example of how you might use Databricks to distill useful information from complex data.
But first, let’s consider four common reasons why the value of data is not fully realised in an organisation:
Separation between data analysis and data context.
Those who have deep data analytic skills – data engineers, statisticians, data scientists – are often in their own specialised area with a business. This area is separated from those who own and understand data assets. Such a separation is reasonable: most BAU data collection streams don’t have a constant demand for advanced analytical work, and often advanced analytical projects require data sourced from a variety of business functions. Unfortunately, success requires strong engagement between those that deeply understand the data and those that deeply understand the analysis. This sort of strong engagement is difficult to moderate in practice.
We’ve seen cases where because of the short-term criticality of BAU work or underappreciation of R&D work, business data owners are unable to appropriately contribute to a project, leaving advanced analytics team members to make do. We’ve seen cases where all issues requiring data owner clarification are expected to be resolved at the start, and continued issues are taken as a sign that the project is failing. We’ve seen cases where business data knowledge resides solely in the minds of a few experts.
Data analysis requires data context. It’s often said “garbage in, garbage out”, but it’s just as true to say “meaningless data in, meaningless insights out”. Databricks improves this picture by encouraging collaboration between data knowledge holders and data analysts, through its shared notebook-style platform.
Difficulty translating analytical work to production workloads.
Investigation and implementation are two different worlds. Investigation requires flexibility, testing different approaches, and putting “what” before “how”. Implementation requires standards, stability, security and integration into systems that have a wider purpose.
A good example of this difficulty is the (still somewhat) ongoing conflict between the use of Python 2 and Python 3. Python 3 has now almost entirely subsumed Python 2 in functionality, speed, consistency and support. However, due to legacy code, platforms and standards within organisations, there are still inducements to use Python 2, even if a problem is better addressed with Python 3. This same gap can also be found in individual Python modules and R packages. A similar gap can be found in organisational support for Power BI Desktop versions. A more profound gap can be seen if entirely different technologies are used by different areas.
This could either lead to substantial overhead for IT infrastructure sections or substantial barriers to adoption of valuable data science projects. PaaS providers offer to maintain the data analysis platform for organisations, enabling emerging algorithms and data analysis techniques to be utilised without additional infrastructure considerations. Additionally, Databricks supports Python, R, SQL and Scala, which cover the major non-proprietary data analysis languages.
Long advanced analysis iterations.
The two previous issues contribute to a third issue: good advanced analyses take time to yield useful results for the business. By the time a problem is scoped, understood, investigated, confronted and solved the problem may have changed shape or been patched by business rules and process changes enough that the full solution implementation is no longer worth it. Improving the communication between data knowledge holders and data analysts and shortening the distance between investigation and implementation mean that the time between problem and solution is shortened.
What this mean for your organisation is that the business will begin to see more benefits of data science. As confidence and acceptance grow so does the potential impact of data science. After all, more ambitious projects require more support from the business.
Data science accepted as a black box.
Data science is difficult, uncertain and broad. This has three implications. Firstly, a certain amount of unsatisfying results must be expected and accepted. Secondly, there is no single defensible pathway for addressing any given problem. Thirdly, no one person or group can understand every possible pathway for generating solutions. Unfortunately, these implications mean that data science practitioners can be in a precarious position justifying their work. Many decision makers can only judge data science by its immediate results, regardless of the unseen value of the work performed. Unseen value may be recognition of data quality issues or appreciation of better opportunities for data value generation.
We don’t believe in this black box view of data science. Data science can be complicated, but its principles and the justifications within a project should be understood by more than just nominal data scientists. This understanding gap is a problem for an organisation’s maturity in the data science space.
Over recent years wide in-roads have been made into this problem with the rise in usage of notebook-style reports. These reports contain blocks of explanatory text, executable code, code results and mathematical formulas. This mix of functions allows data scientists to better expose the narrative behind their investigation of data. Notable examples of this style are Jupyter Notebooks, R Markdown, or Databricks.
Databricks enables collaboration, platform standardisation and process documentation within an advanced analytics project. Ultimately this means a decreased time between problem identification and solution implementation.
Databricks Example: Biometric Data
For demonstrating Databricks, we have an interesting, real data source: the biometrics collected by our watches and smartphones. You probably also have access to this kind of data; we encourage you to test it out for yourself. For Apple products it can be extracted as an XML file and mounted to the Databricks file system. Not sure how to do this? See our previous article.
Specifically, the data we have is from our national manager for technology, Etienne’s watch and smartphone. Our aim is to extract useful insights from this data. The process we will follow (discussed in the subsequent sections are):
- Rationalise the recorded data into an appropriate data structure.
- Transform the data to be useful for its intended purpose.
- Visualise and understand relationships within the data.
- Model these relationships to describe the structure of the data.
Typically, advanced analytics in the business context should not proceed this way. There, a problem or opportunity should be identified first and the model should be in service of this. However here we have the flexibility to decide how we can use the data as we analyse it. This is a blessing and a curse (as we shall see).
Rationalisation
The process of converting the XML data into a dataframe could be overlooked. It’s not terribly exciting. But it does demonstrate the simplicity of parallelisation when using Databricks. Databricks is built over Apache Spark, an engine designed for in-memory parallel data processing. The user doesn’t need to concern themselves how work is parallelised*, just focus on what they need done. Work can be described using Scala, Python, R or SQL. In this case study we’ll be using Python, which interacts with Spark using the PySpark API.
Since we’ve previously mounted our XML biometrics summary, we can simply read it in as a text file. Note that there are ways to parse XML files, but to see what we’re working with a text file is a bit easier.
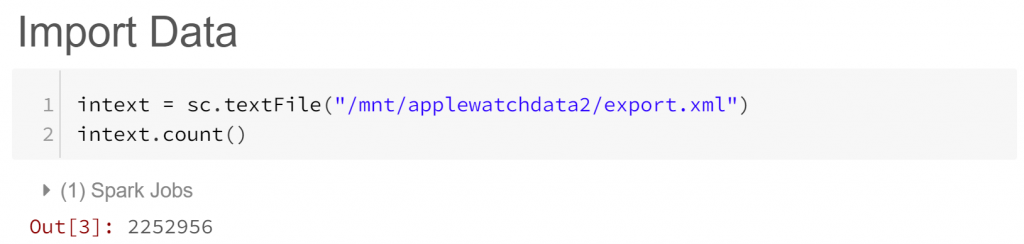
We’ve asked Spark (via sc, a representation of “Spark Context”) to create a Resilient Distributed Dataset (RDD) out of our biometrics text file export.xml. Think of RDDs as Spark’s standard data storage structure, allowing parallel operations across a cluster of machines. In our case our RDD contains 2.25 million lines from export.xml. But what do these lines look like?
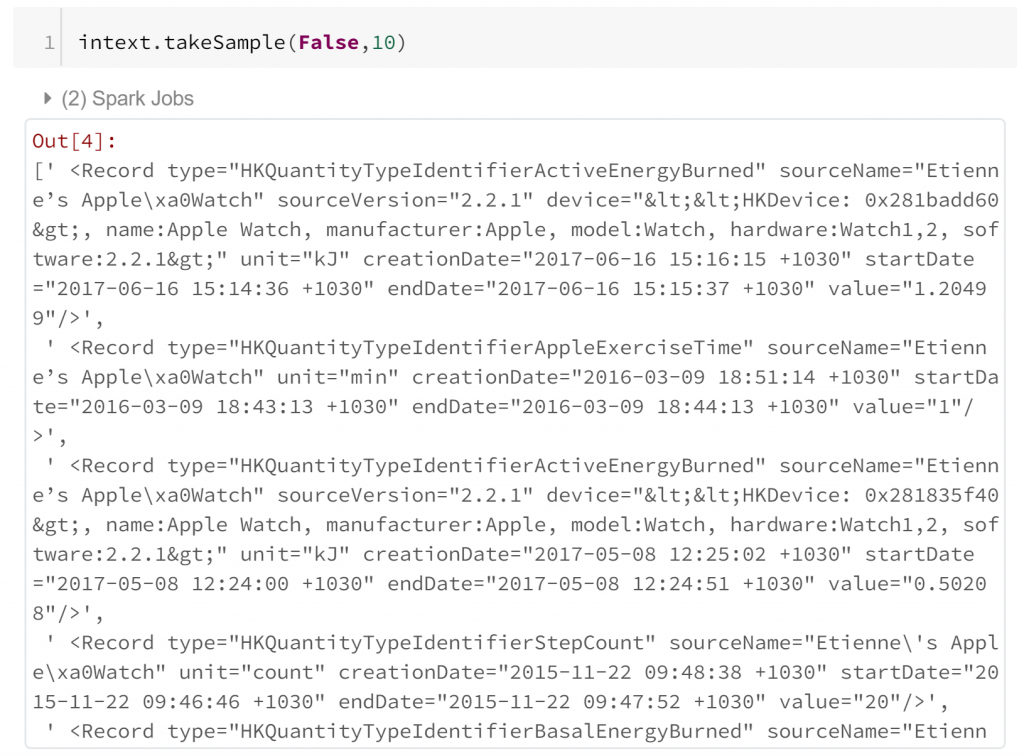
A simple random sample of 10 lines shows that each biometric observation is stored within the attributes of a separate record tag in the XML. This means that extracting this into a tabular format can be quite straight forward. All we need to do is identify record tags and extract their attributes. However, we should probably check that all of our record tags are complete first.

We’ve imported re, a Python module for regular expression matching. Using this we can filter our RDD to find records that begin with “<Record” but are not terminated with “>”. Fortunately, it appears that this is not the case. We can also test for the case where there are multiple records in the same line, but we’ll skip this here. Next we just need to filter our RDD to Record tags.
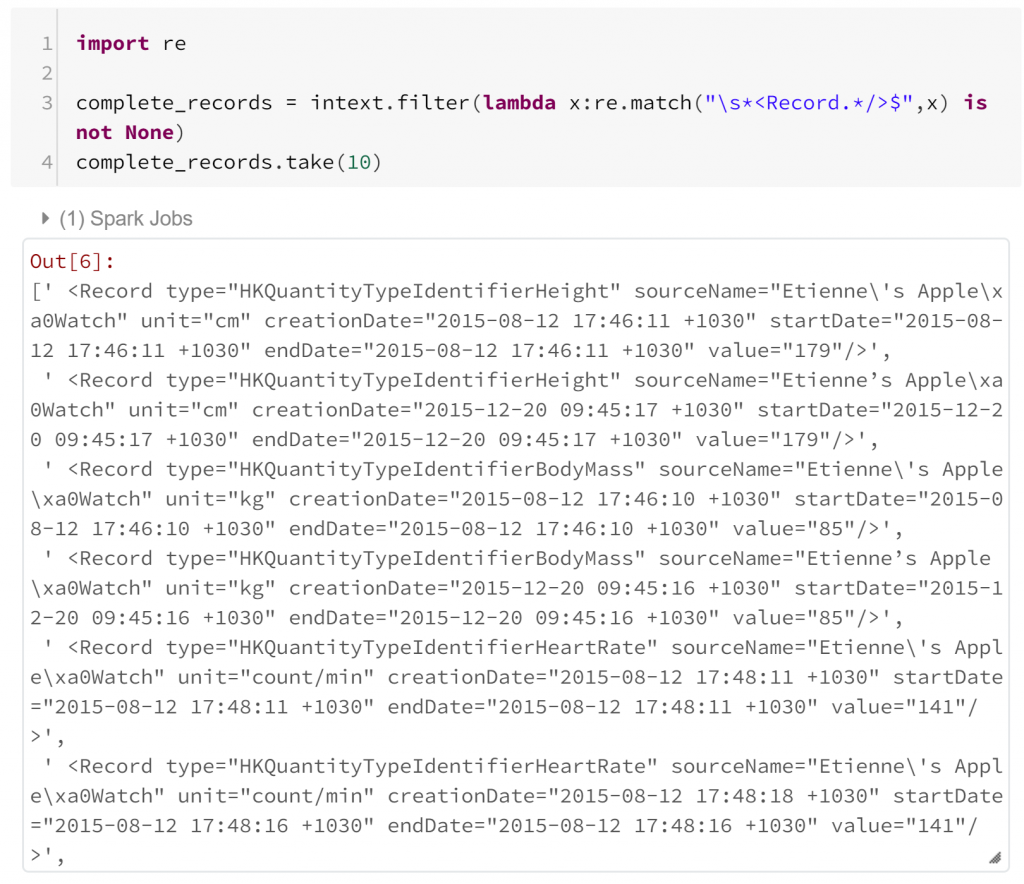
In both of these regular expression checks, I haven’t had to consider how Spark is parallelising these operations. I haven’t had to think any differently from how I would solve this problem in standard Python. I want to check each record has a particular form – so I just import the module I would use normally, and apply it in the Pyspark filter method.
*Okay, not entirely true. Just like in your favourite RDBMS, there are times when the operation of the query engine is important to understand. Also like your favourite RDBMS, you can get away with ignoring the engine most of the time.
Transformation
We already have our records, but each record is represented as a string. We need to extract features: atomic attributes that can be used to compare similar aspects of different records. A record tag includes features as tag attributes. For example, a record may say unit=”cm”. Extracting the individual features from the record strings in our RDD using regular expressions is fairly straightforward. All we need to do is convert each record string into a dictionary (Python’s standard data structure for key-value pairs) with keys representing the feature names and values representing the feature values. I do this in one (long) line by mapping each record to an appropriate dictionary comprehension:
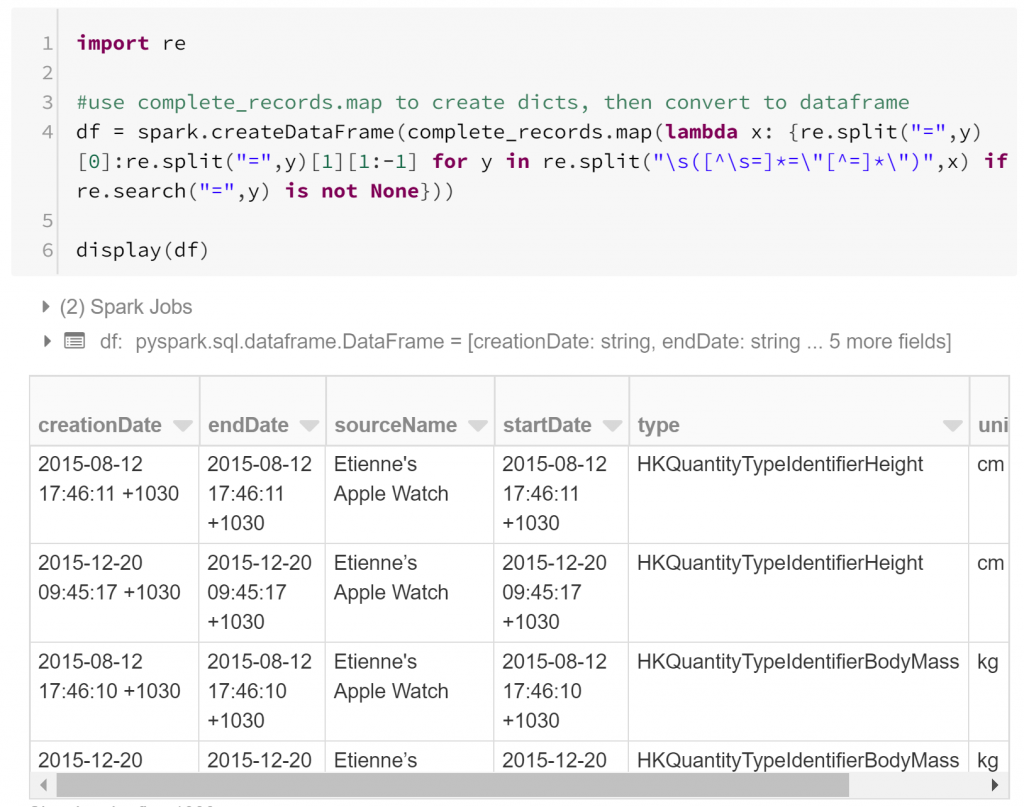
This has converted our RDD into a dataframe – a table-like data structure, composed of columns of fixed datatypes. By and large, the dataframe is the fundamental data structure for data science investigations, inherited from statistical programming. Much of data science is about quantifying associations between features or predictor variables and variables of interest. Modelling such a relationship is typically done by comparing many examples of these variables, and rows of a dataframe are convenient places to store these examples.
The final call to the display function in the above code block is important. This is the default (and powerful) way to view and visualise your data in Databricks. We’ll come back to this later on.
So we have our raw data converted into a dataframe, but we still need to understand the data that actually comprises this table. Databricks is a great platform for this kind of work. It allows iterative, traceable investigations to be performed, shared and modified. This is perfect for understanding data – a process which must be done step-by-step and is often frustrating to document or interpret after the fact.
Firstly in our step-by-step process, all of our data are currently strings. Clearly this is not suitable for some items, but it’s easily fixed.
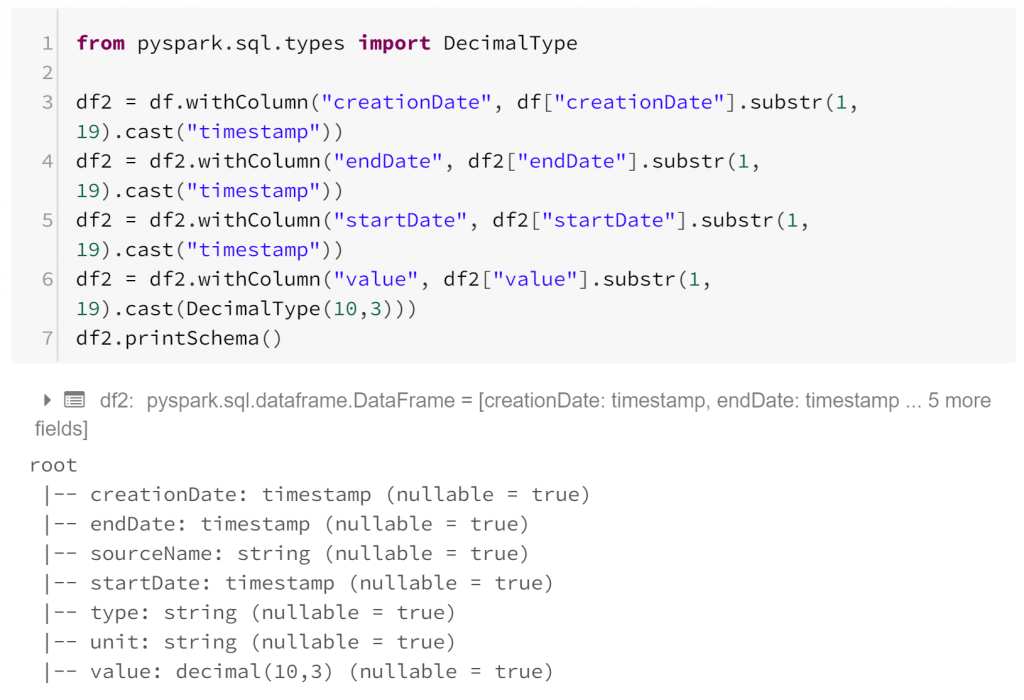
The printSchema method indicates that our dataframe now contains time stamps and decimal values where appropriate. This dataframe has, for each row:
- creationDate: the time the record was written
- startDate: the time the observation began
- endDate: the time the observation ended
- sourceName: the device with which the observation was made
- type: the kind of biometric data observed
- unit: the units in which the observation was measured
- value: the biometric observation itself
Visualisation
So we have a structure for the data, but we haven’t really looked into the substance of the data yet. Questions that we should probably first ask are “what are the kinds of biometric data observed?”, and “how many observations do we have to work with?”. We can answer these with a quick summary. Below we find how many observations exist of each type, and between which dates they were recorded.
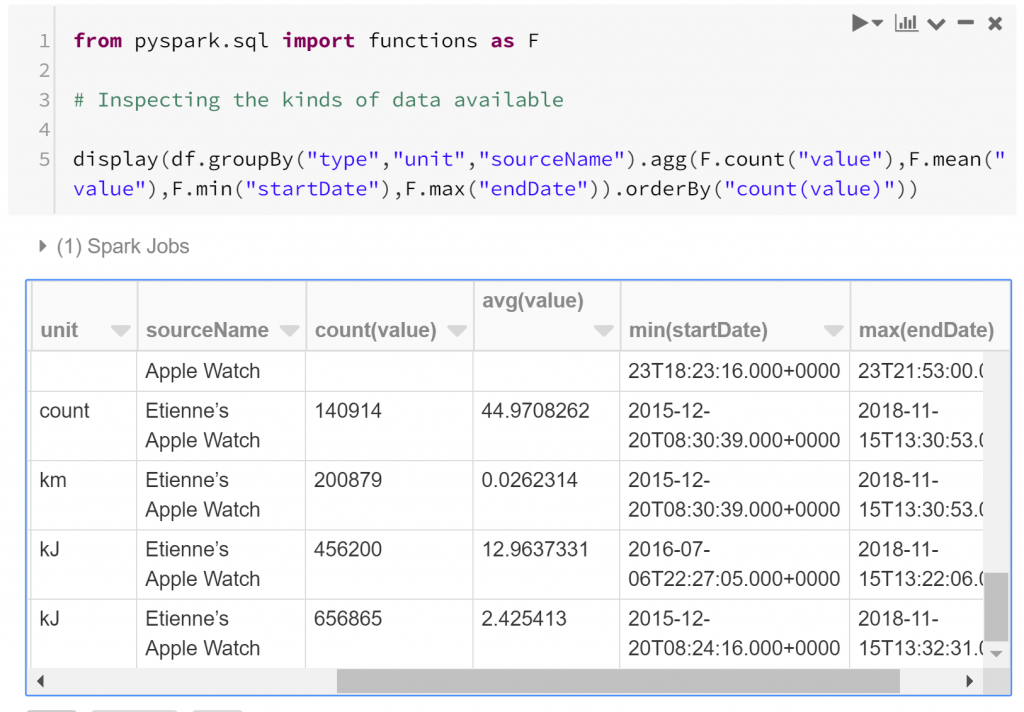
We see that some of the measures of energy burned have the most observations:
- Active Energy Burned has over 650,000 observations between December 2015 and November 2018
- Basal Energy Burned has over 450,000 observations between July 2016 and November 2018
- Distance Walking/Running has over 200,000 observations between December 2015 and November 2018
- Step Count has about 140,000 observations between December 2015 and November 2018
- Heart Rate has about 40,000 observations between December 2015 and November 2017
- Other kinds of observations have less than 30,000 observations
This tells us that the most rich insights are likely to be found by studying distance travelled, step count, heart rate and energy burned. We might prefer to consider observations that are measured (like step count) rather than derived (like energy burned), although it might be an interesting analysis in itself to try to find how these derivations are made.
Let’s begin by looking into how step count might relate to heart rate. Presumably, higher step rates should cause higher heart rates, so let’s see whether this is borne out in the data.
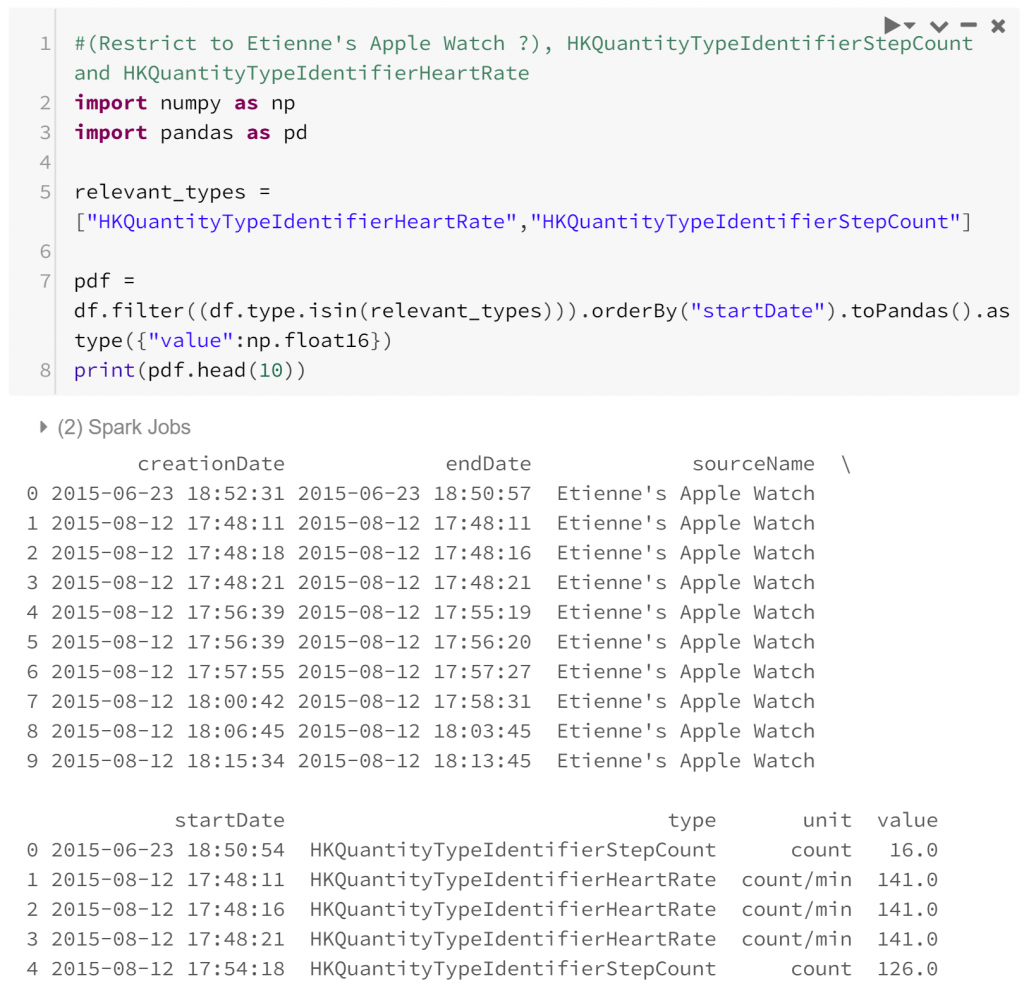
I’ve chosen to convert the data from a Spark dataframe to a Pandas dataframe to take advantage of some of the datetime manipulations available. This is an easy point of confusion for a starter in PySpark: Spark and Pandas dataframes are named the same, but operate differently. Primarily, Spark dataframes are distributed so operate faster with larger datasets. On the other hand, Pandas dataframes are generally more flexible. In this case since we’ve restricted our analysis to a subset of our original data that’s small enough to be confident with a Pandas dataframe.
Actually looking at the data now, one problem appears: the data are not coherent. That is, the two kinds of observations are difficult to compare. This manifests in two ways:
- Heart rate is a point-in-time measurement, while step count is measured across a period of time. This is a similar incoherence to the one in economics surrounding stock and flow variables. To make the two variables comparable we can assume that the step rate is constant across the period of time the step count is measured. As long as the period of time is fairly short this assumption is probably quite reasonable.
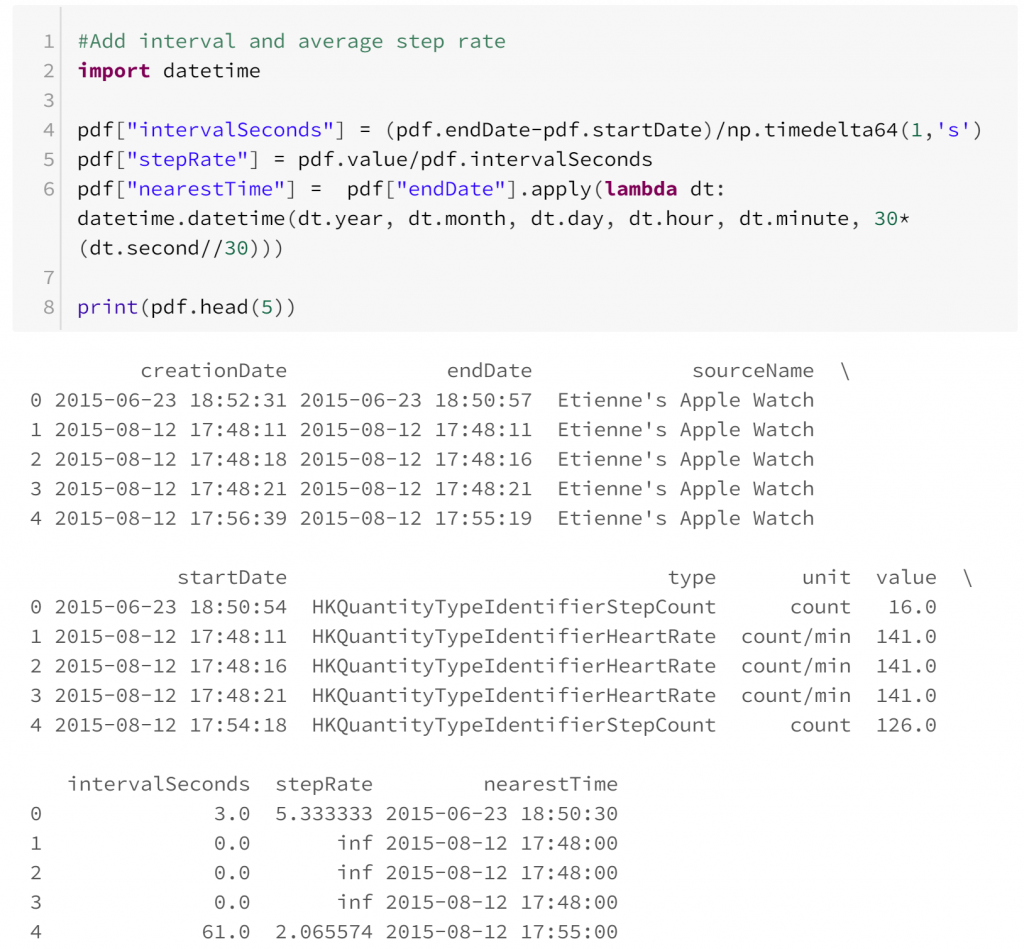
- Heart rate and step count appear to be sampled independently. This means that comparing them is difficult because at times where heart rate is known, step count is not always known, and vice versa. In this case we could assume that both types of observation are sampled independently so we can restrict our comparisons to observations of heart rate and step rate that are reasonably close.
Once we have some observations of heart rate and step rate, we can compare them:
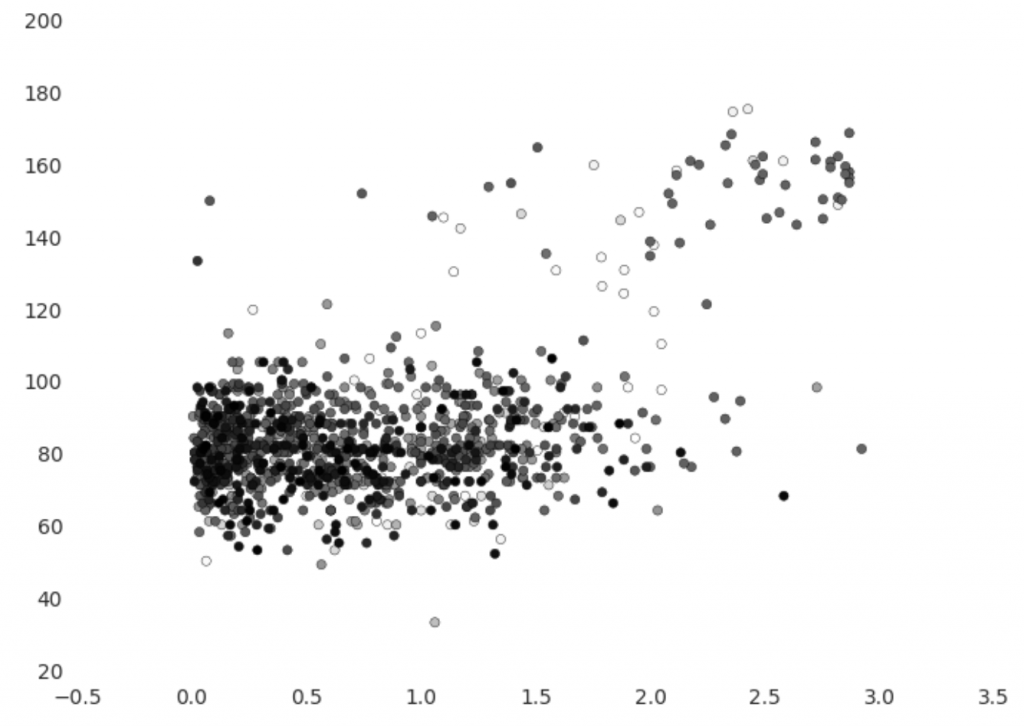
On the vertical axis we have heart rate in beats per minute and on the horizontal axis we have pace in steps per second. Points are coloured so that older points are lighter, which allows us to see if there is an obvious change over time. The graph shows that Etienne’s usual heart rate is about 80 bpm, but when running it increases to between 120 and 180. It’s easy to notice an imbalance between usual heart rate observations and elevated heart rate observations – the former are much more prevalent.
There appears to be at least one clear outlier – the point where heart rate is under 40 bpm. There are also a small amount of observations that have normal heart rate and elevated pace or vice versa – these may be artifacts of our imperfect reconciliation of step count and heart rate. We could feed this back to improve the reconciliation process or re-assess the assumptions we made, which would be particularly useful with subject matter expert input.
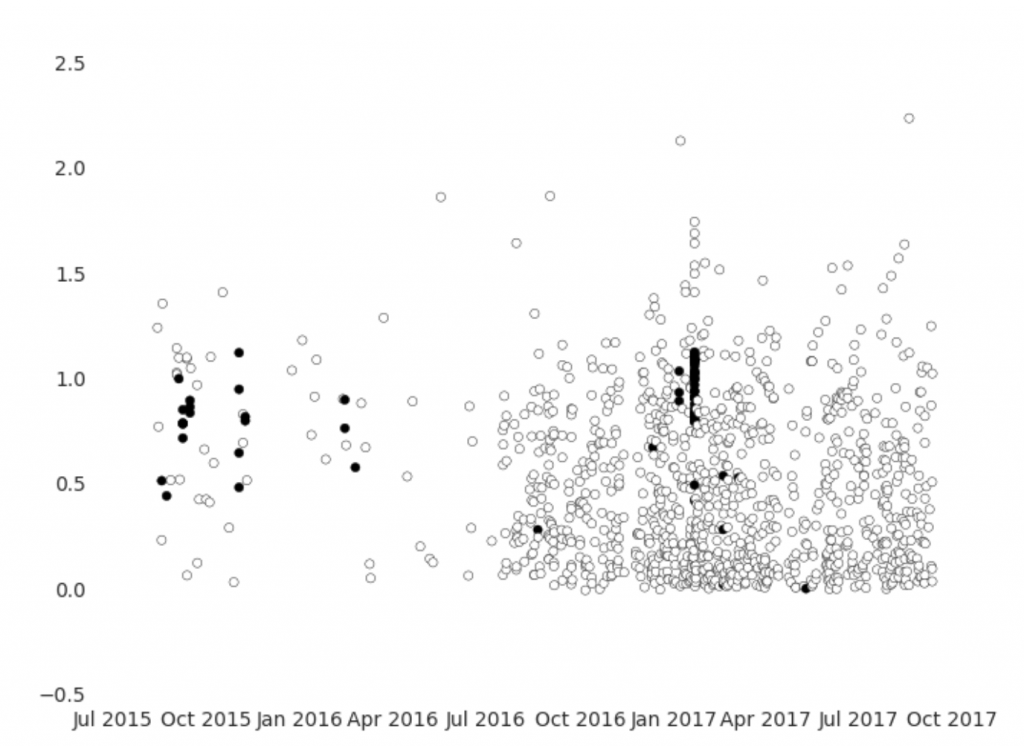
The graph above shows the observations of step rate over time, with black indicating observations that have elevated heart rates. There are a few interesting characteristics – most obviously, observations are far more dense after July 2016. Also, rather alarmingly, there are only a small number of clusters of observations with elevated heart rates, which means that we cannot treat observations as independent. This is often the case for time series data, and it complicates analysis.
We could instead compare the progression of heart rate changes with pace by looking at each cluster of elevated heart rate records as representative of a single exercise events. However, we would be left with very few events. Rather than attempt to clean up the data further, let’s pivot.
Transformation (Iteration 2)
Data doesn’t reveal all of their secrets immediately. Often this means our analyses need to be done in cycles. In our first cycle we’ve learned:
- Data have been collected more completely since mid-2016. Perhaps we should limit our analysis to only the most recent year. This means we should not perhaps attempt to identify long-term changes in the data.
- Heart rate and step rate are difficult to reconcile because they often make observations at different times. It would be better to focus on a single type of biometric.
- There are only a small number of reconcilable recorded periods of elevated heart rate and step rate. Our focus should be on observations where we have more examples to compare.
Instead of step count and heart rate, let’s instead look at patterns in distance travelled by day since 2017. This pivot answers each of the above issues: it is limited to more recent data, it focuses on a single type of biometric data, and it allows us to compare on a daily basis. Mercifully, distance travelled is also one of the most prevalent observations in our dataset.
You’d be right to say that this is a 180 degree pivot. We’re now looking at an entirely different direction. This is an artifact of our lack of a driving business problem, and it’s something you should prepare yourself for too if you commission the analysis of data for the sake of exploration. You may find interesting insights, or you may find problems. But without a guiding issue to address there’s a lot of uncertainty about where your analysis may go.
Stepping down from my soapbox, let’s transform our data. What I want to do is to record the distance travelled in every hourly period from 8am to 10pm since 2017. Into a dataframe “df_x”, I’ve placed all distance travelled data for 2017:

In the above we tackle this in three steps:
- Define a udf (user defined function) which returns the input number if positive or zero otherwise
- Use our udf to iteratively prorate distance travelled biometrics into the whole hour between 8am and 10pm that they fell into, naming these columns “hourTo9”, up to “hourTo22”.
- Aggregate all distances travelled into the day they occurred
This leaves us with rows representing individual calendar days and 14 new columns representing the distance travelled during a hour of the day.
Visualisation (Iteration 2)
This section is not just an exploration of the data, but an exploration of Databricks’ display tool, which allows users to change the output from a code step without re-running the code step. Found at the bottom of every output generated by the display command is a small menu:
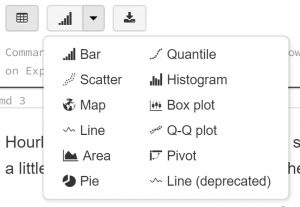
This allows us to view the data contained in the displayed table in a graphical form. For example, choosing “Scatter” gives us a scatterplot of the data, which we can refine using the “Plot Options” dialogue:
We can use these plot options to explore the relationship between the hourly distance travelled variables we’ve created. For example, given a selection of hours (8am to 9am, 11am to 12pm, 2pm to 3pm, 5pm to 6pm, and 8pm to 9pm), we observe the following relationships:
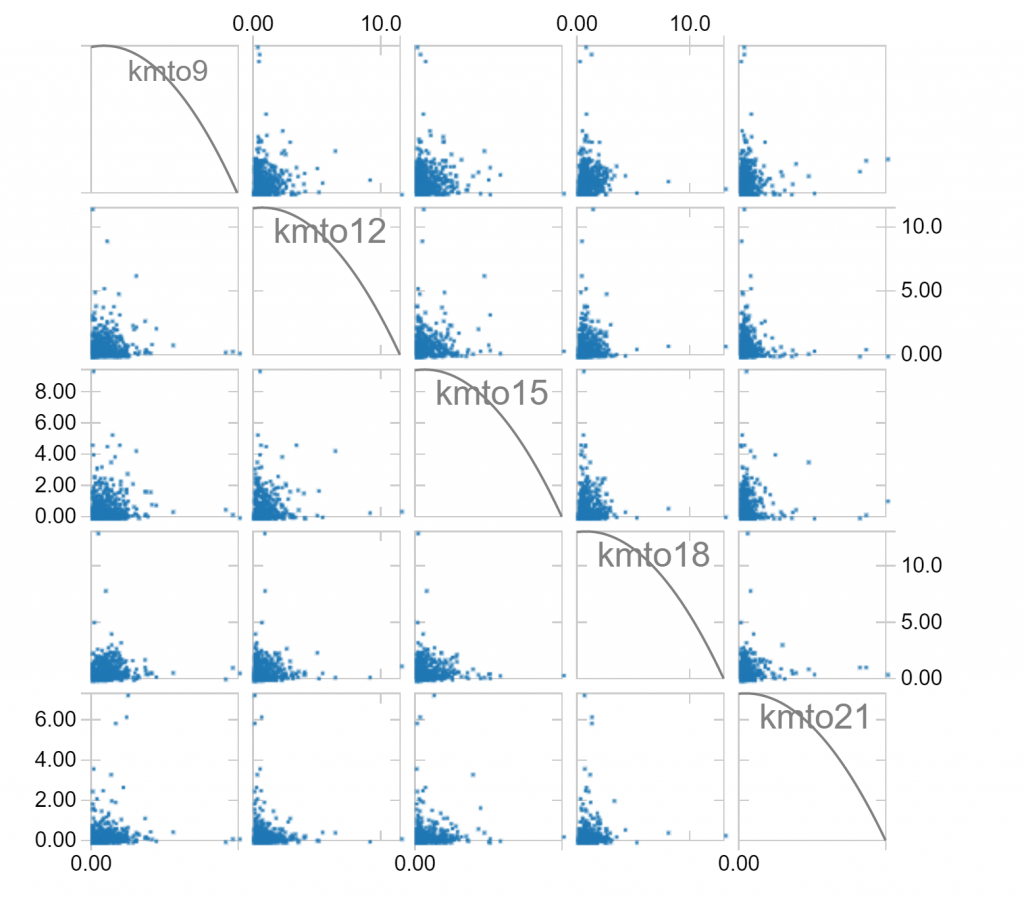
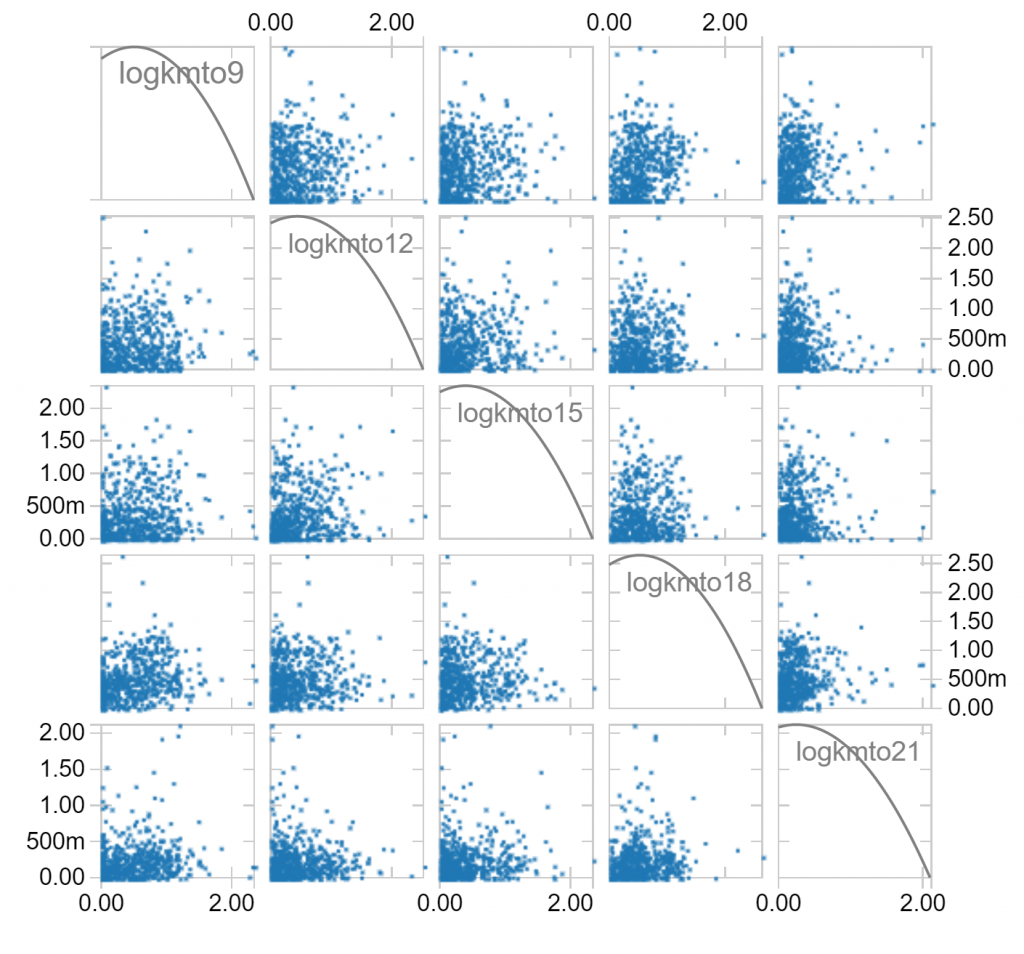
Notice that long distances travelled in one hour of a day makes it less likely that long distances are travelled in other hours. Notice also that there is a fair skew in distances travelled, which is to be expected since the longest distances travelled can’t be balanced by negative distances travelled. We can make a log(1+x) transformation, which compresses large values to hopefully leave us with less skew:
The features we have are in 14 dimensions, so it’s hard to visualise how they might all interact. Instead, let’s use a clustering algorithm to classify the kinds of days in our dataset. Maybe some days are very sedentary, maybe some days involve walking to work, maybe some days include a run – are we able to classify these days?
There are a lot of clustering algorithms at our disposal: hierarchical, nearest-neighbour, various model-based approaches, etc. These perform differently on different kinds of data. I expect that there are certain routines within days that are captured by the data with some random variation: a set jogging route that occurs at roughly the same time on days of exercise, a regular stroll at lunchtime, a fixed route to the local shops to pick up supplies after work. I think it’s reasonable to expect on days where a particular routine is followed, we’ll see some approximately normal error around the average case for that routine. Because of this, we’ll look at using a Gaussian Mixture model to determine our clusters:
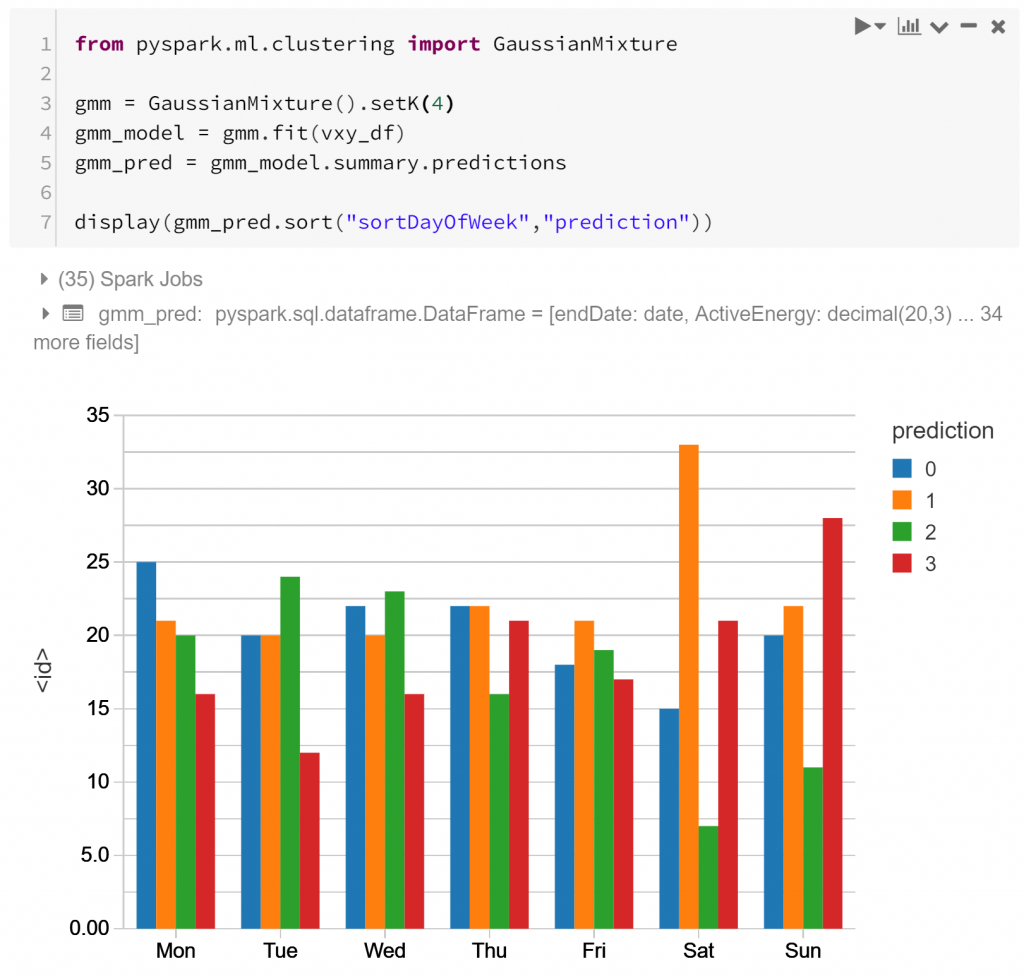
I’ve arbitrarily chosen to cluster into 4 clusters, but we could choose this more rigorously. 4 is enough to show differences between different routines, but not too many for the purpose of demonstration.
The graph above shows the 4 types of routine (labelled as “prediction” 0-3), and their relative frequency for each day of the week. Notably type 1 is much more prevalent on Saturday than other days – as is type 3 for Sunday. Type 2 is much more typical a routine for weekdays, appearing much less on weekends. This indicates that perhaps there is some detectable routine difference between different days of the week. Shocking? Not particularly. But it is affirming to see that the features we’ve derived from the data may capture some of these differences. Let’s look closer.
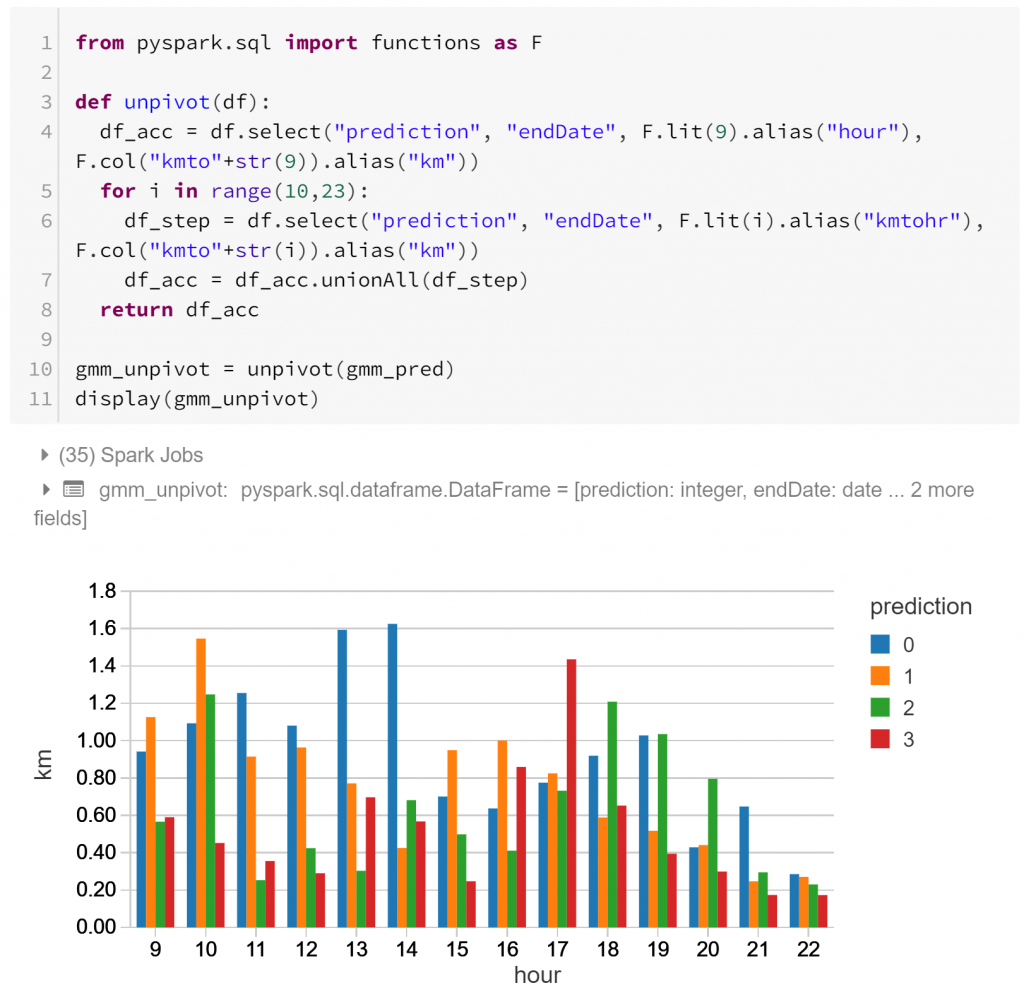
Above we have the actual profiles of the types of daily routines, hour-by-hour. Each routine has different peaks of activity:
- Type 0 has sustained activity throughout the day, with a peak around lunchtime (12pm – 2pm).
- Type 1 has sustained activity during the day with a local minimum around lunchtime, and less activity in the evening.
- Type 2 has little activity during core business hours, and more activity in the morning (8am – 10am) and evening (5pm-7pm)
- Type 3 has a notable afternoon peak (3pm – 6pm) after a less active morning, with another smaller spike around lunchtime.
If you were doing a full analysis you would also be concerned about the variability within and between each of these routine types. This could indicate that more routines are required to describe the data, or that some of the smaller peaks are just attributable to random variation rather than actual characteristics of the routine.
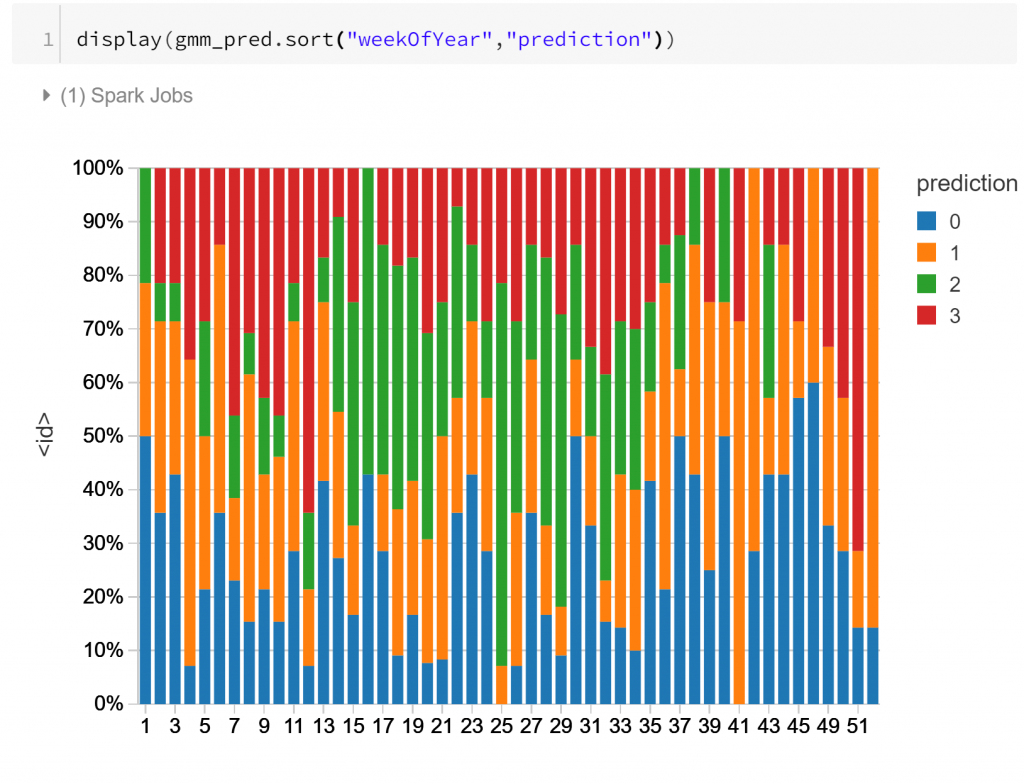
Finally, the visualisation above shows the composition of the daily routines over the course of a year, labelled by week number. The main apparent change through the course of the year is for routine type 2, which is more frequent during cooler months. This concords with what we might suspect: less activity during business hours in cooler, wetter months.
Taken together, perhaps we can use the hourly distance features to predict whether a day is more likely a weekday or a weekend. This model might not seem that useful at first, but it could be interesting to see which weekdays are most like weekends – perhaps these correspond with public holidays or annual leave?
Modelling
Let’s do a quick model to prove that weekends can be classified just with hourly movement data. There are a lot of possible ways to approach this, and a lot of decisions to make and justify. As a demonstrator here we’ll create a single model, but won’t refine it or delve too deeply into it.
Based on the types of routines identified in our cluster analysis, it’s fair to suspect that there may not be a monotonic relationship between the distance travelled in any particular hour and weekend/weekday membership. So rather than using the simplest classification model, logistic regression*, let’s fit a random forest classifier. First, we need to include a label for weekends and weekdays. I choose to call this “label” because by default this is the column name that Pyspark’s machine learning module will expect for classification.
As usual to allow us to check for overfitting, let’s separate the data into a training set and a test set. In this case we have unbalanced classes, so some might want to ensure we’re training on equal numbers of both weekdays and weekends. However, if our training data has the same relative class sizes as the data our model will be generalised to and overall accuracy is important then there isn’t necessarily a problem with unbalanced classes in our training data.
Now let’s prepare for model training. We’ll try a range of different parametrisations of our model, with different numbers of trees, and different numbers of minimum instances per node. Cross-validation is used to identify the best model (where best is based on the BinaryClassificationEvaluator, which uses area under ROC curve by default).

Fitting the model is then simply a matter of applying the cross-validation to our training set:

Finally, we can evaluate how successful our model is.:

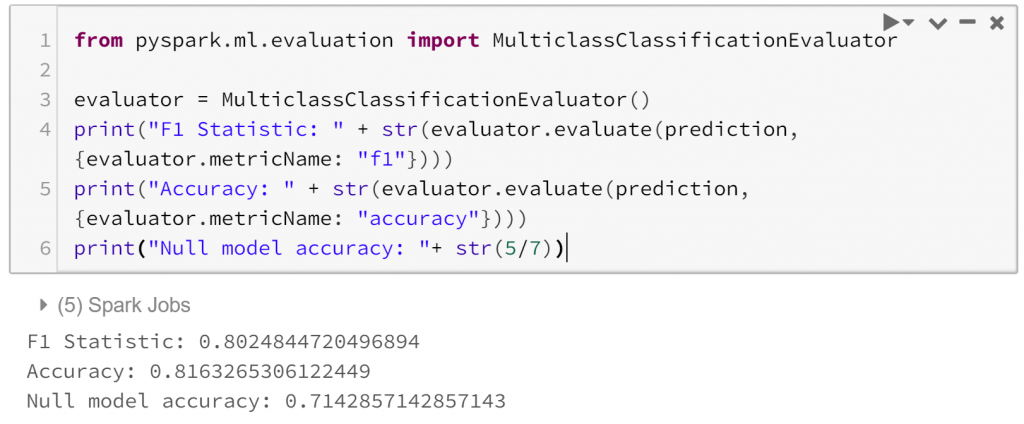
So our model is reasonable on our test data, with a test ROC curve covering 0.86 and an overall accuracy of 0.82, which compares favourably to the accuracy of our null model, which would classify all observations as a weekday and have an accuracy of 0.71. There are many more possible avenues to investigate, even within the narrow path we’ve taken here. This is a curse of exploratory analysis.
*To be fair, logistic regression can capture non-monotonicity as well, but this requires modifying features (perhaps adding polynomial functions of features)
Wrapping Up
Databricks gives us a flexible, collaborative and powerful platform for data science, both exploratory and directed. Here we’ve only managed to scratch the surface, but we have shown some of the features that it offers. We also hope we’ve shown some of the ways it addresses common problems businesses face bringing advanced analysis into their way-of-working. Databricks is proving to be an important tool for advanced data analysis.
